Virtual datasets: Cutting is easier than glueing
To understand why managing km-scale ESM output is challenging, it is first necessary to look at how it is produced. A multi-decadal global simulation at 10 km resolution can easily generate petabytes of output over the course of a single experiment. The large grid sizes and the massive number of both variables and time steps results in tens of thousands of individual files for the model components involved. Each of these files contains only a small part of the overall picture. This explosion in file count makes it extremely difficult to perform any analysis that spans large time or spatial scales. Thousands of separate files must be opened and read — a process that is dominated by heavy input/output (I/O) operations. On top of this, the traditional format, NetCDF, carries a few inherent inefficiencies when used for km-scale climate model output. Each file often contains redundant metadata — for example, repeating coordinate and dimension definitions. This duplication can inflate total storage size by 10% or more, which becomes a serious overhead at Petabyte scale. Worse, to analyze or visualize the data as a single continuous dataset, users must often merge these separate files into one — a process we can think of as “gluing” thousands of puzzle pieces together. This gluing takes considerable time, consumes additional storage, and must be repeated by every researcher who needs access to the same experiment.

Here, virtual means that the data are not duplicated or moved; instead, a metadata definition tells the system where each variable, time step, or spatial chunk is located across all the original files. The metadata are stored once, while the data remain in their original location. Users can then open this virtual dataset through a single interface — the Zarr library — as if it was one big file. And crucially, they can subset it, or “cut” it, to access only the parts they need (for example, one variable, one region, or one decade), without ever having to assemble or read the full dataset first.
This shift from gluing to cutting changes everything. Analysts no longer need to wait for long pre-processing steps or keep local copies of merged datasets. They can query what they need on demand, directly from storage. This saves both time and space. Data managers benefit, too: maintaining a single metadata layer instead of thousands of merged files greatly simplifies updates and ensures consistency across experiments.
This is exactly how EERIE handles km-scale model data. For each simulation experiment, EERIE generates Zarr-based virtual datasets and indexes them in searchable catalogs. These virtual datasets are enriched with valuable metadata such as coordinates, variable attributes, and experiment information. This makes them as analysis-ready as possible. At the same time, they preserve the integrity of the raw model output.
The catalogs, in turn, act as structured gateways: they list all available virtual datasets and include ready-to-use access recipes. With the latter, analysis tools receive exact instructions and connection details on how to open each dataset. This means users can explore, open, and analyze data directly from the catalog without needing any additional information or configuration files.
By following open-source practices, EERIE shares all scripts and configuration templates for generating these virtual datasets openly on GitLab. This transparency allows other projects to adopt the same approach.
The result is a more efficient way of working with high-resolution climate data. Instead of spending time gluing files together, researchers can now focus on cutting straight to insight — exploring, comparing, and analyzing vast model outputs through a unified, streamlined interface. Feel invited to visit our documentation on the easy.gems platform — it will guide you through the world of EERIE data.
Cloudify: A cloud storage emulator
Bringing data to cloud storage (like we all know from services from Google or Amazon) is, in theory, the simplest solution to disseminate and increase the impact of Earth System Model (ESM) output for both data providers and data producers. Hosting data on cloud storage is easy and flexible, especially when contrasted with traditional on-premise server solutions. Recognizing this, DKRZ supports the community by preparing and testing a new S3 cloud storage service.
However, to make ESM output efficiently accessible via cloud storage, it must first be stored in a cloud-optimized format.
In the case of EERIE, having already taken the first step with virtual datasets, storing high-resolution model output as Zarr in cloud storage might seem like the obvious next move — after all, Zarr is specifically designed for cloud storage. Yet, in practice, several obstacles make this more complicated than it seems.
First, institutional cloud resources are often limited; even at DKRZ. Second, converting legacy formats like NetCDF or GRIB into fully optimized Zarr datasets is both time-consuming and computationally expensive. It requires careful coordination between data producers and users. Finally, most model outputs remain stored locally on HPC systems. Migrating them to the cloud would rather duplicate the data and waste valuable storage space.
To overcome these challenges, the DKRZ team developed Cloudify (Wachsmann et al., 2025), a cloud storage emulator built on xpublish, an open-source framework that serves datasets over web protocols with Zarr-compatible endpoints. Instead of physically copying data to the cloud, Cloudify makes local datasets behave as if they were already in the cloud. It exposes the data through a RestAPI that look and act like cloud storage interfaces, even though the data remain on local disks in HPC systems. This approach allows researchers and tools to interact with the data without the cost or complexity of full data migration.
First Cloudify was developed on a virtual machine called eerie.cloud (Wachsmann et al. 2026, paper in preparation) as an experimental service to offer open and standardized Zarr access to all EERIE simulations. What began as a small proof of concept quickly turned into a powerful bridge between HPC and cloud-based workflows. Researchers could now explore Petabytes of high-resolution climate data using familiar cloud tools, while the data itself never left the disk storage. Over time, eerie.cloud evolved into a shared, production-grade service — adopted for projects such as NextGEMs, Orcestra, and DYAMOND — and is now growing into DKRZ’s dedicated “km-scale cloud.” Nearly 6 Petabyte of EERIE simulation data (in memory size) are made available through the eerie.cloud (see Table 1) , which in total exceeds the S3 size DKRZ could allocate projects as a fair share.
For scientists, this means seamless, near-instant access to complex model output without downloads or file merging. For data providers, it offers a scalable, efficient way to make their results discoverable and reusable. And for the broader community, eerie.cloud shows how thoughtful design can turn existing HPC infrastructure into a truly open, cloud-like environment — bringing cutting-edge climate simulations within everyone’s reach.
| Project | Datasets | Source format | Total size in memory [Terabyte] | Resolution [km] |
| ORCESTRA | 104 | Zarr | 30 | 1 |
| ERA5 | 7 | GRIB | 373 | 28 |
| Cosmo-rea | 12 | NetCDF | 118 | 12 |
| nextGEMS | 35 | Mixed | 3058 | 5 |
| EERIE | 386 | Mixed | 5895 | 10 |
| Dyamond | 16 | Mixed | 232 | 2 |
| Sum | 560 | 9706 |
Table 1: Project datasets available via the km-scale-cloud in 07/2025.
Catalogs: Discover DKRZ data stores
After making km-scale model output easier to access and cloud-ready through virtual datasets and Cloudify, discoverability still remains as the last challenge.
Even if the data are technically available, it’s easy to feel lost. Which experiments are available? Where are they stored? How can they be opened? With hundreds of experiments, model versions, variables, and resolutions, the landscape of high-resolution simulations can feel like a vast, uncharted archive. Finding the right dataset quickly and confidently is as important as accessing it efficiently.
This is where catalogs come into play. In EERIE’s data workflow, catalogs serve as the maps that guide users through the data landscape. They describe each dataset, explain how to open it, and connect related experiments into a navigable structure.
EERIE’s data pipeline initially leveraged Intake, a lightweight, cataloging tool that works smoothly with Python tools for data analysis. With just a few lines — often less than ten — data providers define a dataset’s location, structure, and access method, linking directly to our virtual datasets in the EERIE intake catalog (Wachsmann et al, 2024). Once defined, these catalogs can be queried directly from Python scripts or Jupyter notebooks. A single function call can load a full km-scale dataset, enabling users to move from metadata to analysis in seconds. This approach made it easy for EERIE scientists to prototype workflows, test new diagnostics, or automate batch analyses without manual data handling.
However, over time, limitations of Intake became visible. Intake provides no graphical interface. Nor are newer versions backward-compatible. And it cannot interoperate easily with tools other than Python. To make our catalogs more open, durable, and widely usable,a more universal approach that could bridge languages, platforms, and communities is needed.
That’s when EERIE joined forces with DKRZ’s broader effort to adopt STAC — the SpatioTemporal Asset Catalog standard. Originally developed for Earth observation data, STAC has since become a flexible, cross-domain metadata framework for describing and indexing any dataset with spatial or temporal structure. It’s both human- and machine-readable, accessible through graphical interfaces, and discoverable via APIs. DKRZ co-develops a STAC-based infrastructure within projects like WarmWorld, building a foundation that supports discovery, search, and access across many scientific domains. The DKRZ STAC world can already be explored through https://discover.dkrz.de
Within EERIE, the team laid the foundation for efficient future STAC indexing of km-scale ESM output. We created STAC item templates specifically tailored for our virtual datasets. Each dataset comes with rich metadata — including experiment descriptions, coordinate information, direct access URLs. In Figure 1, the STAC-browser view on such an item is illustrated.
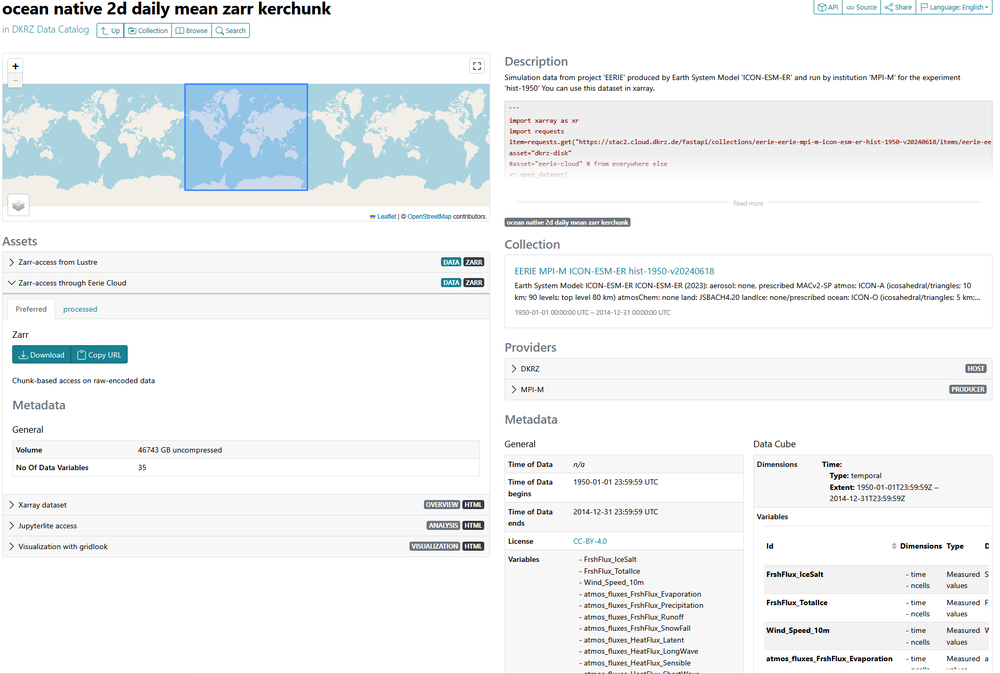
Figure 1: An EERIE STAC item for an ICON dataset that includes ocean variables for daily frequency, displayed in the web browser. Users can quickly see how to access the dataset and what it contains.
It also provides code examples showing how to open the data programmatically. Additionally, each STAC item includes links to open-source applications and visualization tools (see figure 2) that use the data, making it not only discoverable but immediately usable. Afterwards these items were grouped into STAC collections, for example, a collection for the hist-1950 experiment run with the ESM ICON.
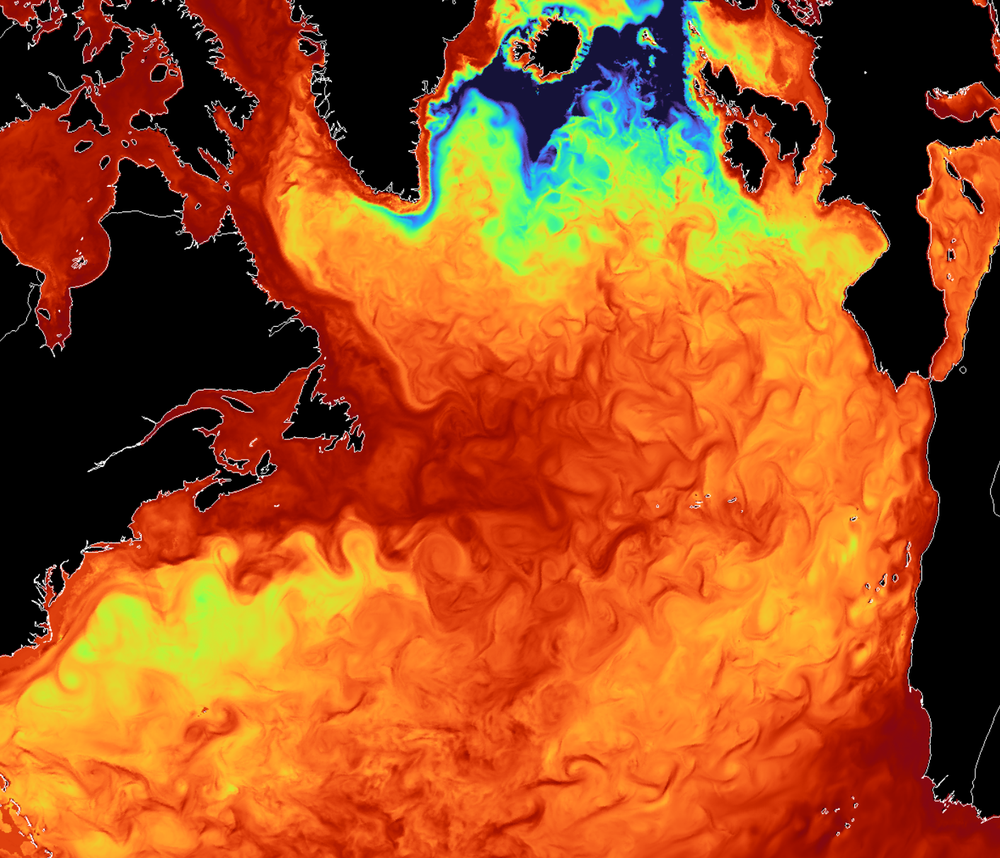
Figure 2: The mixed layer depth over the North Atlantic in the EERIE ICON hist-1950 simulation with a resolution of ~5km visualised with gridlook. If you can download >100 Megabyte through your network, you can create this plot by clicking this weblink. It will render on your GPU a time step of the virtual dataset accessed live through the eerie.cloud.
Visitors to eerie.cloud are greeted by a browsable interface of the full EERIE main collection — a structured, living catalog that connects every experiment, dataset, and tool in one place. Behind the scenes, this catalog is continuously updated as new simulations finish, new experiments are added, and new metadata are generated from the EERIE pipeline.
The future of climate data management
Together, virtual datasets, Cloudify, and catalogs form the backbone of EERIE’s open data ecosystem. The virtual datasets make km-scale climate model data usable; Cloudify makes it accessible; and the catalogs make it discoverable. This pipeline transforms what was once a fragmented collection of files into a coherent, searchable, and open resource for climate science.
For researchers working with the EERIE data ecosystem means spending less time preprocessing or downloading data — and more time doing science. EERIE doesn’t just store data — it turns it into knowledge waiting to be found.

This work has received funding from the Swiss State Secretariat for Education, Research and Innovation (SERI) under contract #22.00366.
This work was funded by UK Research and Innovation (UKRI) under the UK government’s Horizon Europe funding guarantee (grant number 10057890, 10049639, 10040510, 10040984).
References:
Wachsmann, Fabian; Heil, Angelika; Wickramage, Chathurika; Polkova, Iuliia; Thiemann, Hannes; Modali, Kameswarrao; Lammert, Andrea; Peters-von Gehlen, Karsten; Kindermann, Stephan (2025). Technical Overview of Cloudify - An Improved Emulator of Cloud-Optimized Earth System Model Output. World Data Center for Climate (WDCC) at DKRZ. https://doi.org/10.35095/WDCC/Overview_Cloudify
Wachsmann, F., Matthias Aengenheyster, Wickramage, C., Seddon, J., Nikolay Koldunov, & Ziemen, F. (2024). Data access to EERIE ESM output via intake catalogues: Phase1 plus Hackathon simulations (v1.0.0). Zenodo. https://doi.org/10.5281/zenodo.14243677
Authors:
- Fabian Wachsmann, department: Data Management German Climate Computing Center (DKRZ),
- Chathurika Wickramage, department: Data Management, German Climate Computing Center (DKRZ)
- EERIE and Data Management team at DKRZ